
Theme 3: Enabling Algorithms
The Enabling Algorithms theme facilitates the use and advancement of Algorithms across all ACEMS themes. Enabling Algorithms are characterized as enhancing (enabling) computation across a range of applications.
This includes (a) developing new and enhanced algorithms to solve a variety of methodological and applied problems, (b) presenting theoretical and empirical proofs and evidence that the algorithms work as claimed, and (c) providing clear descriptions of the algorithms and easy to use code. ACEMS researchers have been at the forefront of building on currently available methods as well as creating new algorithms at the cutting edge of research. During 2020, the Enabling Algorithm researchers made significant advances in key algorithms that are used to analyse and solve questions in a wide variety of fields, including statistical physics, brain physiology, computational intelligence, forensics, and cancer research.
The Numbers
Researchers
Cross-node collaboration groups
Industry and international collaboration projects
People
Nigel Bean, Kevin Burrage, Tim Garoni, Rob Hyndman, Robert Kohn*, Dirk Kroese*, Kerrie Mengersen, Scott Sisson, Kate Smith-Miles, Ian Turner*, Matt Wand
(* Theme Leaders)
Azam Asanjarani, Davaatseren Baatar, Peter Ballard, Boris Beranger, Andrew Black, Pamela Burrage, Julian Caley, Jessica Cameron, Chris Carter, Nathan Clisby, Andrea Collevecchio, Dianne Cook, Paul Corry, Tiangang Cui, Hans De Sterck, Simon Denman, Christopher Drovandi, Troy Farell, Denzil Fiebig, David Frazier, Claudia Grazian, Jens Grimm, Shovanaur Haque, Markus Hegland, Kate Helmstedt, Wang Jin, Sevvandi Kandanaarachchi, Jonathan Keith, Bonsoo Koo, Brodie Lawson, Catherine Leigh, Jessica Liebig, Benoit Liquet, Fawang Liu, Gael Martin, James McGree, Ross McVinish, Matthew Moores, Mario Andres Muñoz Acosta, Giang Nguyen, John Ormerod, Anastasios Panagiotelis, Guoqi Qian, Rachael Quill, Matias Quiroz, Adam Rohrlach, Fred Roosta, Joshua Ross, Thomas Taimre, Priyanga Talagala, Thiyanga Talagala, Emi Tanaka, Duncan Taylor, Nicholas Tierney, Minh Ngoc Tran, Radislav Vaisman, Mattias Villani, Paul Wu, Hongbo Xie, Nan Ye
Doan Khue Dung (KD) Dang, Xuhui Fan, Megan Farquhar, Puwasala Gamakumara, David Gunawan, Sarat Moka, Pablo Montero Manso, Steven Psaltis, Robert Salomone, Matthieu Simon, Chris van der Heide, David Warne, Eric Zhou
Ziwen An, Igor Balnozan, Edward Barker, Mark Baxendale, Joshua Bon, Chuntao Chen, Vincent Chin, Rixon Crane, Hung Dao, Laurence Davies, Mohammed Javad Davoudabadi, Dilishiya De Silva, Vektor Dewanto, Zhe Ding, Ethan Goan, Thomas Goodwin, Sayani Gupta, Adam Hamilton, Virginia He, Jacinta Holloway-Brown, Shuwen Hu, Wathsala Karunarathne, Shamika Prasadini Kekulthotuwage Don, Aline Kunnel, Stuart Lee, Angus Lewis, Dan Li, Yang Lui, Amalan Mahendran, Hayden Moffat, Ryan Moseni, Abrahim Nasrawi, Aaron Snoswell, Susumu Tsuchida, James Walker, Riley Whebell, John Worrall, Yu Yang, Hui Yao, James Yu
Hugh Andersen, Sarah Belet, Imke Botha, Anny Francis, Valentina di Marco, Ryan Kelly, Stephanie Kobakian, Dylan Morris, Jon Peppinck, Jacob Priddle
Lachlan Bennett, Callum Gray, Mitchell O’Sullivan, Jonathan Wilton
Mitchell O’Hara-Wild
Department of Agriculture and Fisheries (QDAF)
Dr Huan Du (CentraleSupélec, Université Paris-Saclay, France), Antonietta Mira (University of Lugano, Switzerland), David Nott (National University of Singapore), Professor Patrick Perré (CentraleSupélec, Université Paris-Saclay, France), Zongze Yang (Northwestern Polytechnical University, China)
Key Achievements
In 2020, ACEMS researchers continued developing novel methodologies and improving existing methods to solve a range of problems.
Researchers at UQ wrote papers, developing and analysing new algorithms in a wide range of fields, including numerical linear algebra, computational intelligence, forensics, and cancer research. Some of the highlights are outlined below.
AI Fred Roosta and his students developed new and improved algorithms for second-order optimization methods for neural networks. The findings were published in prestigious journals and conferences such as ACM/IEEE Supercomputing Conference (SC20), ICML, and SIAM Journal on Optimization.
Research Fellows Sarat Moka and Christopher van der Heide along with AI Radislav Vaisman and CI Dirk Kroese developed and analyzed several new Monte Carlo algorithms for the estimation and simulation of spatial phenomena.
The results were published in esteemed journals such as Bernoulli and Nature Communications. One of the research projects focused on chromosome arm aneuploidies (CAAs), or copy number imbalances, which are pervasive in cancers. How they affect cancer development, prognosis and treatment remains largely unknown. CAA profiles of 23,427 tumours were analysed, identifying aspects of tumour evolution including probable orders in which CAAs occur and CAAs predicting tissue-specific metastasis. The researchers found that CAAs can predict cancer prognosis, shape tumour evolution, metastasis and drug response, and may advance precision oncology.
AI Nan Ye made significant advances in Deep Learning algorithms, more specifically Nesterov acceleration of alternating least squares, published in top-quality proceedings such as IJCAI and ICLR.
CI Kroese and AIs Vaisman, and Thomas Taimre enabled the PDF of their 2019 book Data Science and Machine Learning: Mathematical and Statistical Methods to be downloaded for free from the ACEMS website. In 2020, the page received 4,316 page views which is significantly higher than any page on the ACEMS website.
Students working in this theme also had impact via a number of achievements, Aaron Snoswell won first place and People’s Choice Award at the Faculty of Engineering, Architecture and Information Technology 3 Minute Thesis Final at UQ, he was also interviewed on Triple J radio about his research. Jonathan Wilton was awarded an AMSI research scholarship, Susumu Tsuchida completed their PhD and Lachlan Bennett and Callum Gray completed Honours.
CI Ian Turner, in collaboration with Professor Patrick Perré and Dr Huan Du from CentraleSupélec, Université Paris-Saclay in France, developed a generalised fractional transport model to simulate the growth of a colony of wood-rot fungus, Postia placenta. We believe this was the first time that fractional calculus has been applied to model fungal colonisation and the work was published in Communications in Nonlinear Science and Numerical Simulation.
CI Turner, AI Fawang Liu, PhD students and research collaborators from China continued their work on the development of efficient, stable and convergent computational algorithms for solving fractional partial differential equations. A notable application of our work was developed in collaboration with visiting researcher Zongze Yang, where a fractional Bloch-Torrey model was used for simulating the transport of cerebrospinal fluid (CSF) in the ventricles of the brain. The work was published in the Journal of Computational Physics.
CI Turner and Research Fellow Steven Psaltis continued their research with the Department of Agriculture and Fisheries (QDAF). In their recent paper accepted for publication in the Annals of Forest Science, they reported their collaborative research on the development of a novel non-destructive method for predicting the modulus of elasticity (MOE) of logs using measurements taken from cores extracted from discs cut from these logs. In an extension to this work, they showed how increment cores can provide improved predictive capabilities of the MOE of sawn boards. This work has been submitted for publication in the Annals of Forest Science. Work also continued with QDAF on investigating the impact of continuous drying on key production and performance criteria of engineered wood structural elements. Their numerical simulations show excellent agreement with the experimental drying data.
Research Fellow Psaltis continued his work with collaborators at the University of Oxford on modelling of lithium batteries. This project began through the ACEMS International Mobility Program. They have developed novel multiscale reduced-order models of the electrical and thermal behaviour in spirally-wound (or jelly-roll) cells. This work has been submitted for publication in SIAM Journal on Applied Mathematics and is available on arXiv.
Researchers used advanced likelihood-free Bayesian methods to reveal new insights in biology and epidemiology. One paper published in PLOS Computational Biology developed a new stochastic model for transmission of banana bunchy top virus on a banana plantation. The model calibrated with a likelihood-free method can be used to monitor and forecast various disease management strategies. The paper was led by undergraduate Engineering student Abhishek Varghese, and involved AI Chris Drovandi and CI Kerrie Mengersen from ACEMS, and international collaborator Antonietta Mira (University of Lugano, Switzerland). Their work attracted media interest including this news article.
In another relevant paper, AI Drovandi and co-workers used approximate Bayesian computation to estimate a novel stochastic model of COVID-19 transmission. They applied the method/model to more than 150 countries to assess the global response to the early stages of the COVID-19 pandemic. They found that countries imposing strict containment measures early in the epidemic fared better and that broader testing is required earlier in the epidemic. The paper was led by Research Fellow David Warne, and involved AI Drovandi, CI Mengersen and national/international collaborators.
AI Drovandi’s research group has continued to advance Bayesian synthetic likelihood (BSL) for estimating intractable likelihood models. PhD student (now completed) Ziwen An led a paper (published in Statistics and Computing) for making BSL more robust to one of its key assumptions. The method is more robust and retains the advantages of the original BSL method. The paper was published with international collaborator David Nott (National University of Singapore).
Jacob Priddle (completed ACEMS MPhil student) wrote a paper using transformations to massively increase the computational efficiency of BSL whilst losing little accuracy. This paper is still under review but seems on the pathway to publication. The paper involves ACEMS members CIs Ian Turner, Scott Sisson and AI David Frazier. It’s a great example of a successful cross node ACEMS collaboration.
CI Robert Kohn and his collaborators (AI Minh-Ngoc Tran, AI David Gunawan) developed particle-based methodology to tackle complex random effects models and applied it to greatly improve estimation for the linear ballistic accumulator model in cognitive psychology. The methodology is quite general, however, and is suitable for other evidence accumulation models as well as diverse applications in other disciplines. Several application papers in cognitive psychology based on this methodology have been published. In a cross-node collaboration, CI Kohn, together with AI’s Matias Quiroz, Mattias Villani, Minh Ngoc Tran and Robert Salomone show how to use a spectral based approach to carry out subsampling to accelerate Markov chain Monte Carlo (MCMC) computation in stationary time series.
Advances were made in the efficient sampling of spatial random processes, with applications to statistical mechanics.
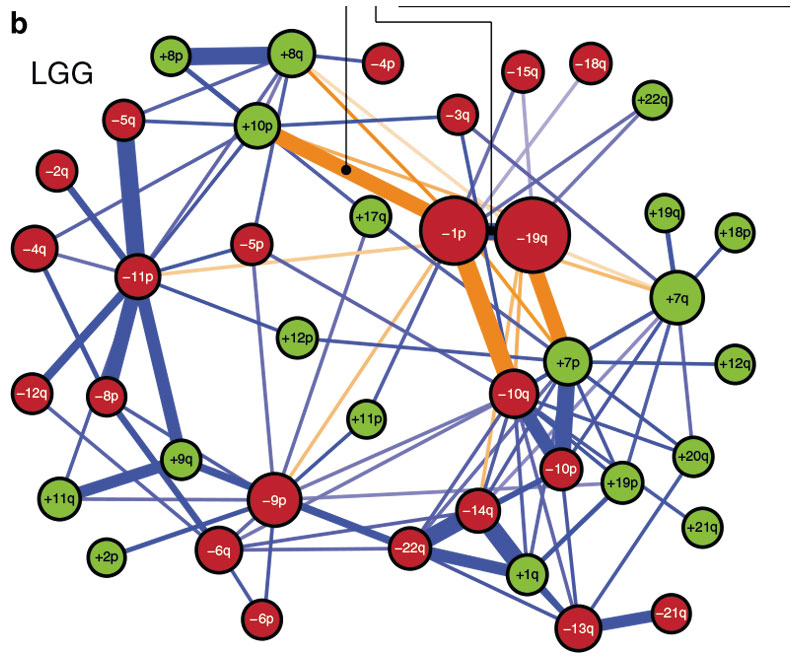
New algorithms were developed to better understand how chromosome abnormalities shape the evolution of tumours.
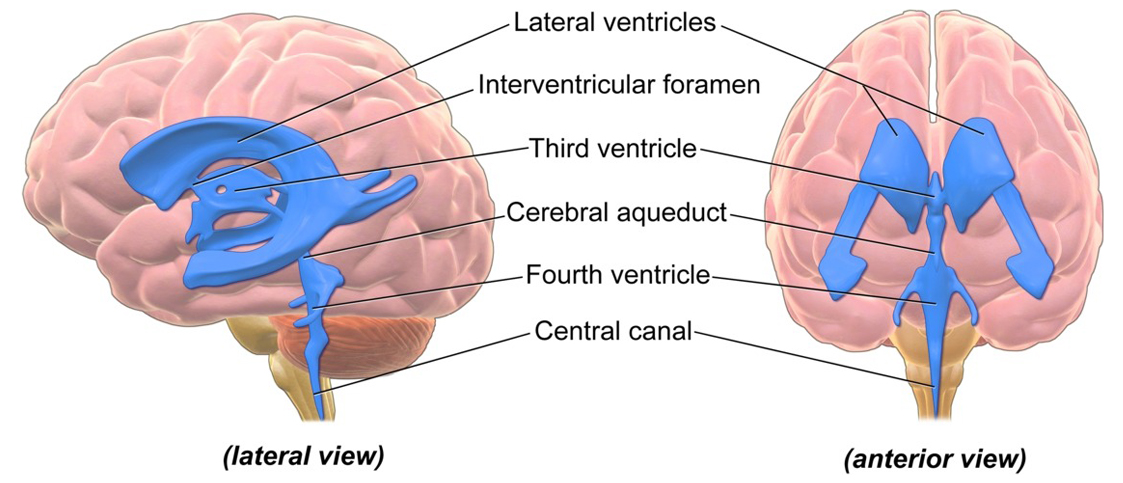
Ventricles of the brain. Ventricles in the blue part (lateral and anterior views).
The mesh of the domain Ω (unit, mm).
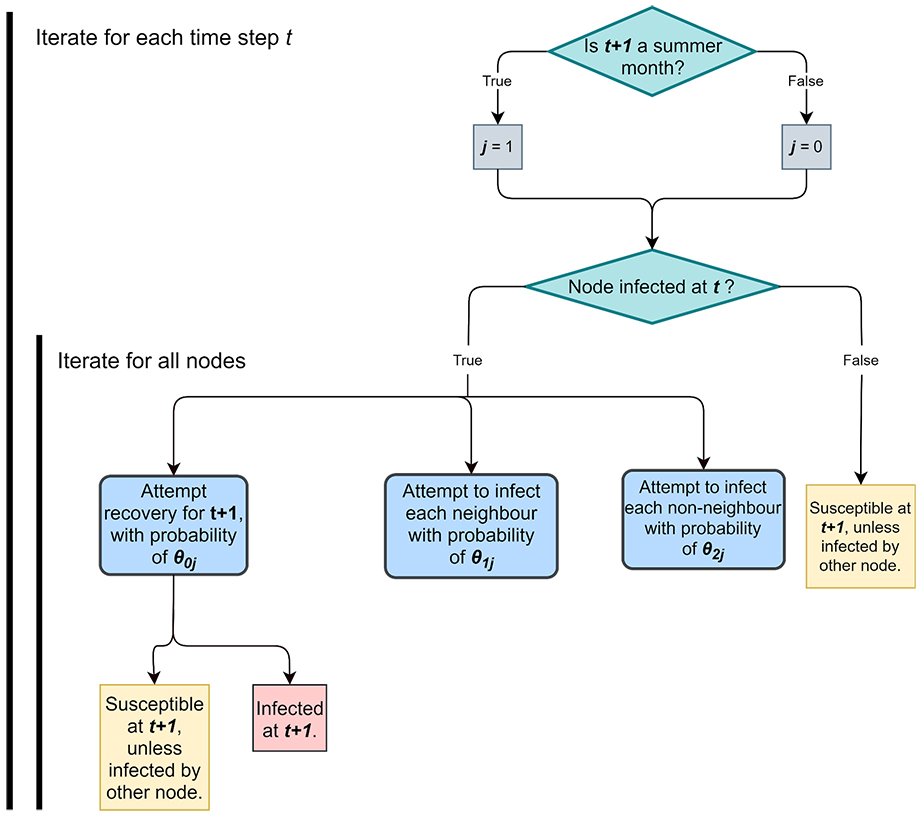
Flow-chart describing banana bunchy top virus model behaviour for each time step t.
Plans for 2021
Plans for 2021 are already well underway, and the following is a brief summary of plans for ACEMS researchers working in the Enabling Algorithms theme:
- Keep pushing the boundaries of Data Science and Machine Learning research, by working on:
- Parallel and distributed learning algorithms (UQ)
- Monte Carlo methods for complex data structures (UQ)
- More robust density tempering and Data Annealing (UNSW)
- Improved Variational Bayes approaches to inference and prediction in time series (UNSW)
- Conduct calibration of continuous drying simulations using approximate Bayesian computation methods.
- Submission of related paper (QUT)
- Establish a new project with QDAF (QUT)
- Modify and extend the existing drying model for simulating water ingress (QUT)
- Extend the water ingress model to simulate mass timber panels composed of many layers of timber (QUT)
- Calibrate the water ingress model to experimental data provided by QDAF (QUT)
- Conduct large-scale simulations of mass timber panels exposed to environmental conditions